A research team has completed a high-quality chromosome-scale assembly of the Chuanhonghua 1 safflower genome. This work sheds light on the genetic underpinnings of crucial traits like linoleic acid (LA) and hydroxysafflor yellow A (HSYA) production. It sets a new precedent for crop improvement and functional genomics studies.
Safflower (Carthamus tinctorius L.), a plant known for its vibrant flowers and nutrient-rich seeds, has been cultivated for thousands of years in the fertile crescent region. It is highly regarded for its oil, rich in LA; and its flowers, which contain HSYA, substances with wide-ranging industrial and medicinal applications. Despite its value, genetic research on safflower has been constrained by the limited understanding of its genome.
A study appearing in Horticulture Research elucidates the genetic underpinnings of safflower, thus paving the way for targeted breeding strategies.
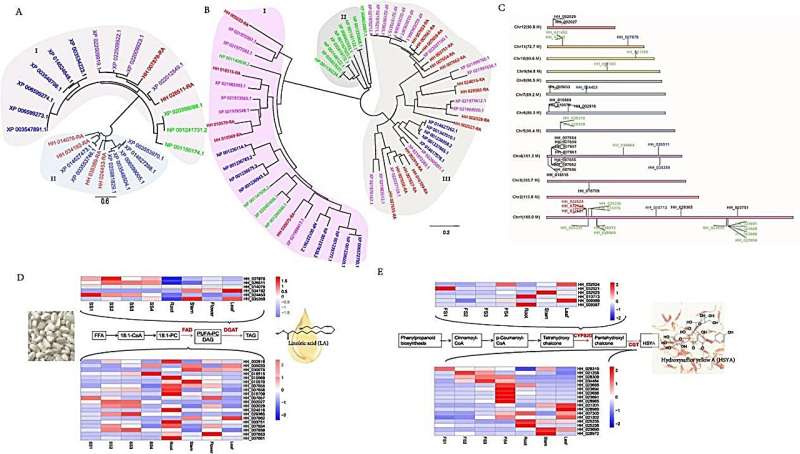
The research team employed an integrated sequencing strategy combining Illumina, Oxford Nanopore, and Hi-C technologies to achieve a high-quality genome assembly of 1.17 Gb with contigs assigned to 12 chromosomes, reflecting about 100-fold coverage relative to the estimated genome size. This assembly facilitated the identification of a recent whole-genome duplication event, underpinning the extensive genomic rearrangements and evolutionary history of safflower, corroborated by phylogenetic analysis using 274 single-copy genes across 12 species, and a detailed examination of gene family expansion and contraction.
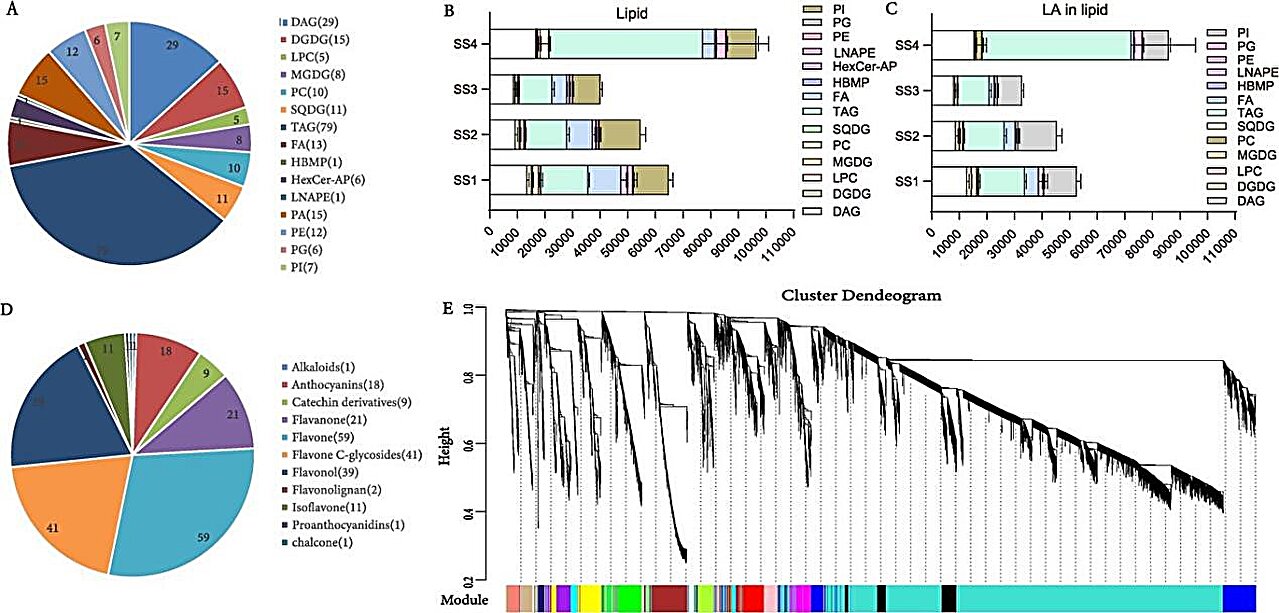
Metabolomic and transcriptomic profiling across different developmental stages of seeds and flowers revealed a comprehensive lipidome with a predominant presence of triacylglycerols (TAGs) and a diverse flavonoid metabolite spectrum, respectively, highlighting the biosynthetic pathways of key compounds such as LA and HSYA. Through genomic analysis, the researchers predicted 39,809 protein-coding genes, with substantial annotation against public databases, and identified specific gene families—diacylglycerol acyltransferase (DGAT) and fatty acid desaturases (FADs) for LA biosynthesis, and CYP and CGT for HSYA biosynthesis—indicating their pivotal roles in these metabolic pathways.
Notably, the re-sequencing of 220 safflower lines yielded 7,402,693 high-quality SNPs, facilitating a genome-wide association study (GWAS) that pinpointed significant SNPs associated with agronomic traits, particularly oil content and flower color. This GWAS analysis, along with the functional verification of the candidate gene HH_034464 (CtCGT1), implicated in HSYA biosynthesis, underscored the potential of molecular markers in enhancing breeding programs for desired traits. The functional assays confirmed CtCGT1’s role in the glycosylation of flavonoids, crucial for HSYA production, thereby shedding light on the genetic underpinnings of key metabolic traits in safflower.
Dr. John Smith, an expert in plant genomics, praised the achievement, stating, “This comprehensive genome assembly is a milestone in safflower research. It not only enhances our understanding of plant evolution but also provides a rich resource for identifying genes responsible for key agricultural and medicinal traits.”
By providing a foundation for the molecular breeding of safflower, this research offers the potential to enhance crop resilience, nutritional content, and therapeutic efficacy. Looking forward, the challenge lies in leveraging this vast genomic resource to develop superior safflower varieties that meet the demands of a changing global climate and growing population.
The chromosome-scale genome assembly of the Chuanhonghua 1 safflower represents a critical step forward in the genetic exploration of this valuable crop. This research not only opens new avenues for safflower functional gene mining and breeding but also has broader implications for the agricultural and pharmaceutical industries.
More information:
Jiang Chen et al, Whole-genome and genome-wide association studies improve key agricultural traits of safflower for industrial and medicinal use, Horticulture Research (2023). DOI: 10.1093/hr/uhad197
Citation:
Unveiling the genetic blueprint of safflower (2024, April 24)
retrieved 24 April 2024
from https://phys.org/news/2024-04-unveiling-genetic-blueprint-safflower.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.