The perception of taste is one of the most important senses and helps us identify beneficial foods and avoid harmful substances. For instance, our fondness for sweet and savory foods results from our need to consume carbohydrates and proteins. Given their importance as an evolutionary trait, researchers around the world are investigating how taste receptors originated and evolved over a period of time. Obtaining these insights into the feeding behavior of organisms can help them paint a picture of the history of life on Earth.
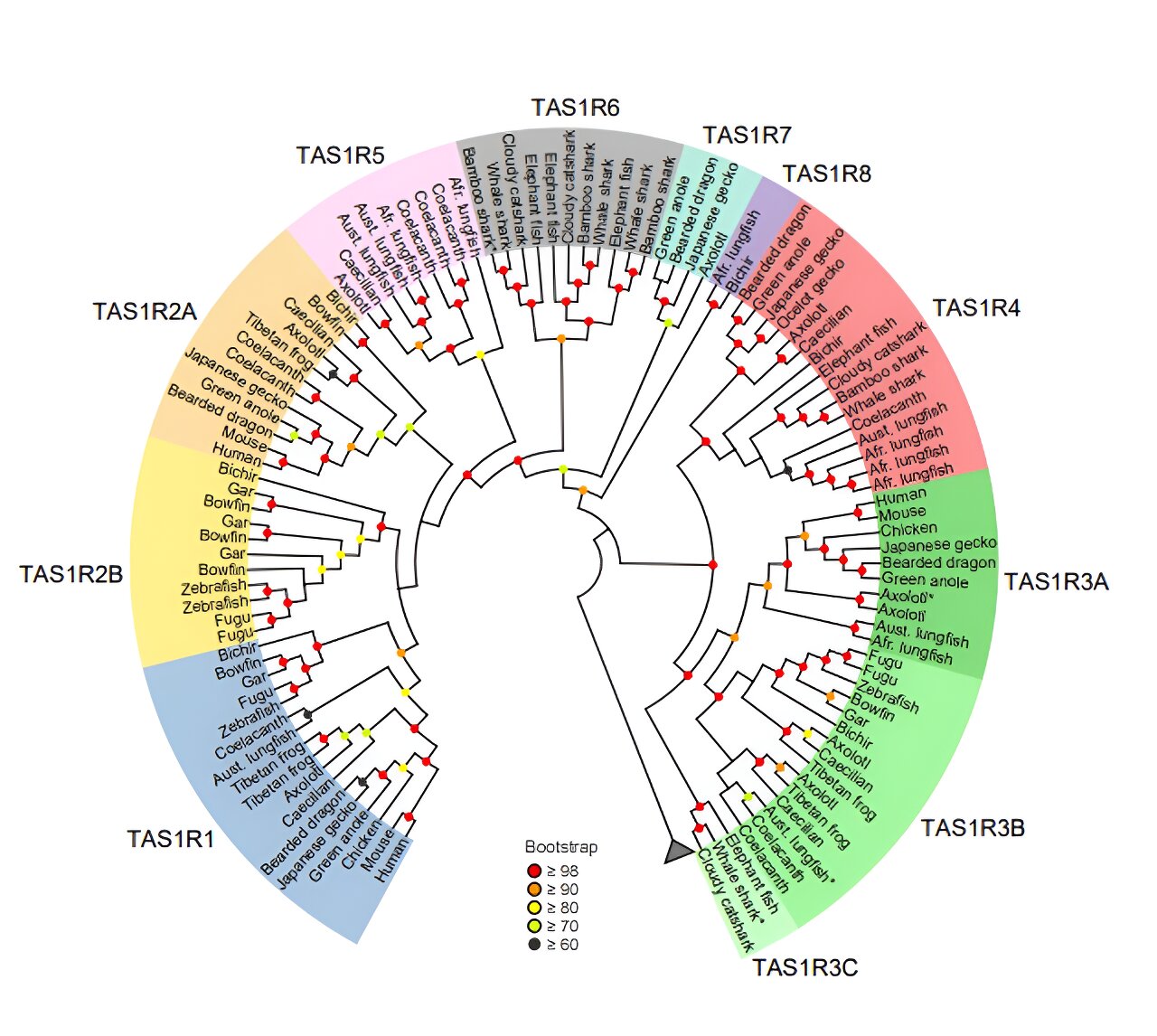
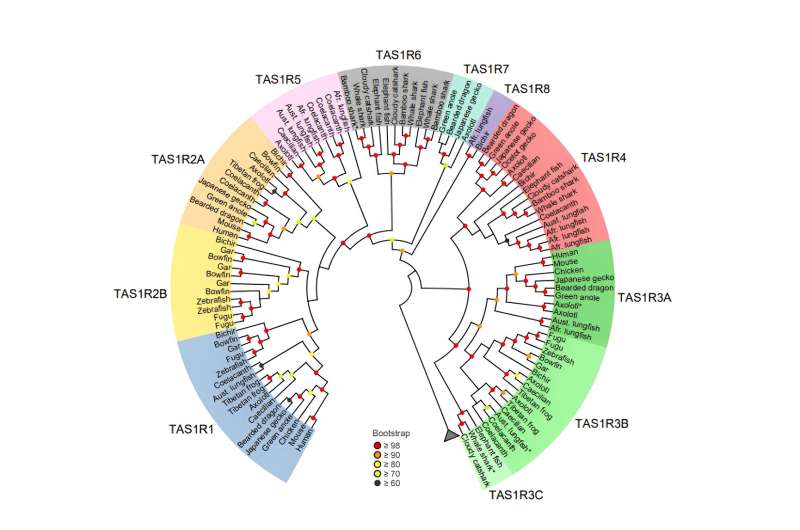
One of the important tastes in our taste palette is umami, or the savory taste, which is associated with proteins that form a vital part of the diets of many organisms. The taste receptor type 1 (T1R) detects sweet and umami tastes among mammals. This taste receptor is encoded by the TAS1R, a family of genes, including TAS1R1, TAS1R2, and TAS1R3, and comes from a common ancestor of bony vertebrates.
However, this gene pattern is not observed in coelacanth and cartilaginous fishes, where “taxonomically unplaced” TAS1R genes have been identified, suggesting an incomplete understanding of the evolutionary history of taste receptors.
Now, however, a research team led by Associate Professor Hidenori Nishihara from Kindai University and Professor Yoshiro Ishimaru from Meiji University, Japan, have identified five new, previously undiscovered groups within the TAS1R family. This discovery is a result of a genome-wide survey of jawed vertebrates including all major fish groups.
The study, published in Nature Ecology & Evolution on December 13, 2023, includes the contributions of Senior Assistant Professor Yasuka Toda from Meiji University, Professor Masataka Okabe from The Jikei University School of Medicine, Professor Shigehiro Kuraku from the National Institute of Genetics, and Project Associate Professor Shinji Okada from The University of Tokyo.
“Our study revealed that as compared to most modern vertebrates, the vertebrate ancestor possessed more T1Rs. These findings challenge the paradigm that only three T1R family members have been retained during evolution,” says Prof. Nishihara.
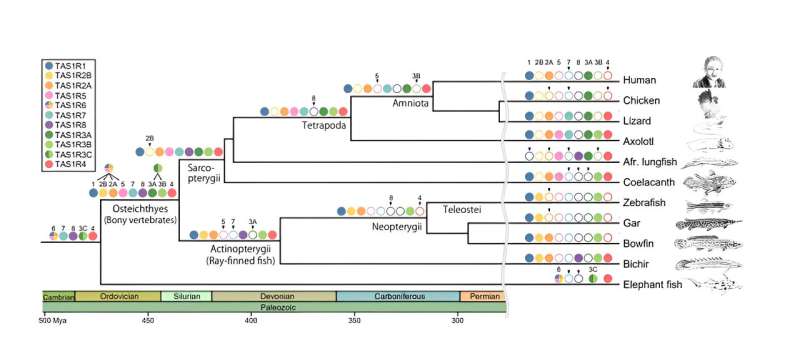
The novel taste receptor genes, named TAS1R4, TAS1R5, TAS1R6, TAS1R7, and TAS1R8 by the researchers, were categorized based on their distribution among species with a common ancestor. The researchers found TAS1R4 genes to be present in lizards, axolotl, lungfishes, coelacanth, bichir, and cartilaginous fishes, but absent in mammals, birds, crocodilians, turtles, and teleost fishes. In addition, axolotl, lungfishes, and coelacanth were found to have TAS1R5.
The researchers observed a close evolutionary relationship between TAS1R5, TAS1R1, and TAS1R2, indicating a shared ancestry between these genes. The cartilaginous fishes possess TAS1R6 exclusively. Notably, the researchers found that TAS1R6 evolved from the same ancestral gene that led to TAS1R1, TAS1R2, and TAS1R5 genes. While axolotl and lizards possess TAS1R7, bichir and lungfishes possess TAS1R8. The researchers determined that these two genes originated in the common ancestor of jawed vertebrates.
In addition to these new genes, the study revealed diversity in the existing TAS1R genes. For instance, they found that TAS1R3 of bony vertebrates could be divided into TAS1R3A and TAS1R3B. TAS1R3A was present in tetrapods and lungfishes, while TAS1R3B was identified in amphibians, lungfishes, coelacanths, and ray-finned fishes. Additionally, the genome survey found TAS1R2 to have diversified into two distinct groups (TAS1R2A and TAS1R2B), challenging the conventional idea that TAS1R2 forms a single gene group.
“We found that the TAS1R phylogenetic tree comprises of a total of 11 TAS1R clades, revealing an unexpected gene diversity,” adds Prof. Nishihara.
The findings also suggest that the first TAS1R gene appeared in jawed vertebrates around 615–473 million years ago. The gene then underwent several duplications to produce nine taste receptor genes (TAS1R1,2A, 2B, 3A, 3B, 4, 5, 7, and 8) in the common ancestor of bony vertebrates. Over time, some of these genes were lost in different lineages, with mammals and teleosts retaining only three TAS1Rs (TAS1R1, TAS1R2A, and TAS1R3A in mammals).
In addition to shedding light on the evolutionary history, the findings also have practical applications. Explaining these to us, Prof. Nishihara says, “These findings make it easier for us to deduce the taste preferences of diverse vertebrates. This, in turn, can have potential applications such as the development of pet foods and attractants tailored to the preferences of fish, amphibians, and reptiles.”
More information:
Hidenori Nishihara et al, A vertebrate-wide catalogue of T1R receptors reveals diversity in taste perception, Nature Ecology & Evolution (2023). DOI: 10.1038/s41559-023-02258-8
Provided by
Kindai University
Citation:
Unraveling the evolutionary origins of umami and sweet taste preferences (2023, December 29)
retrieved 30 December 2023
from https://phys.org/news/2023-12-unraveling-evolutionary-umami-sweet.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.