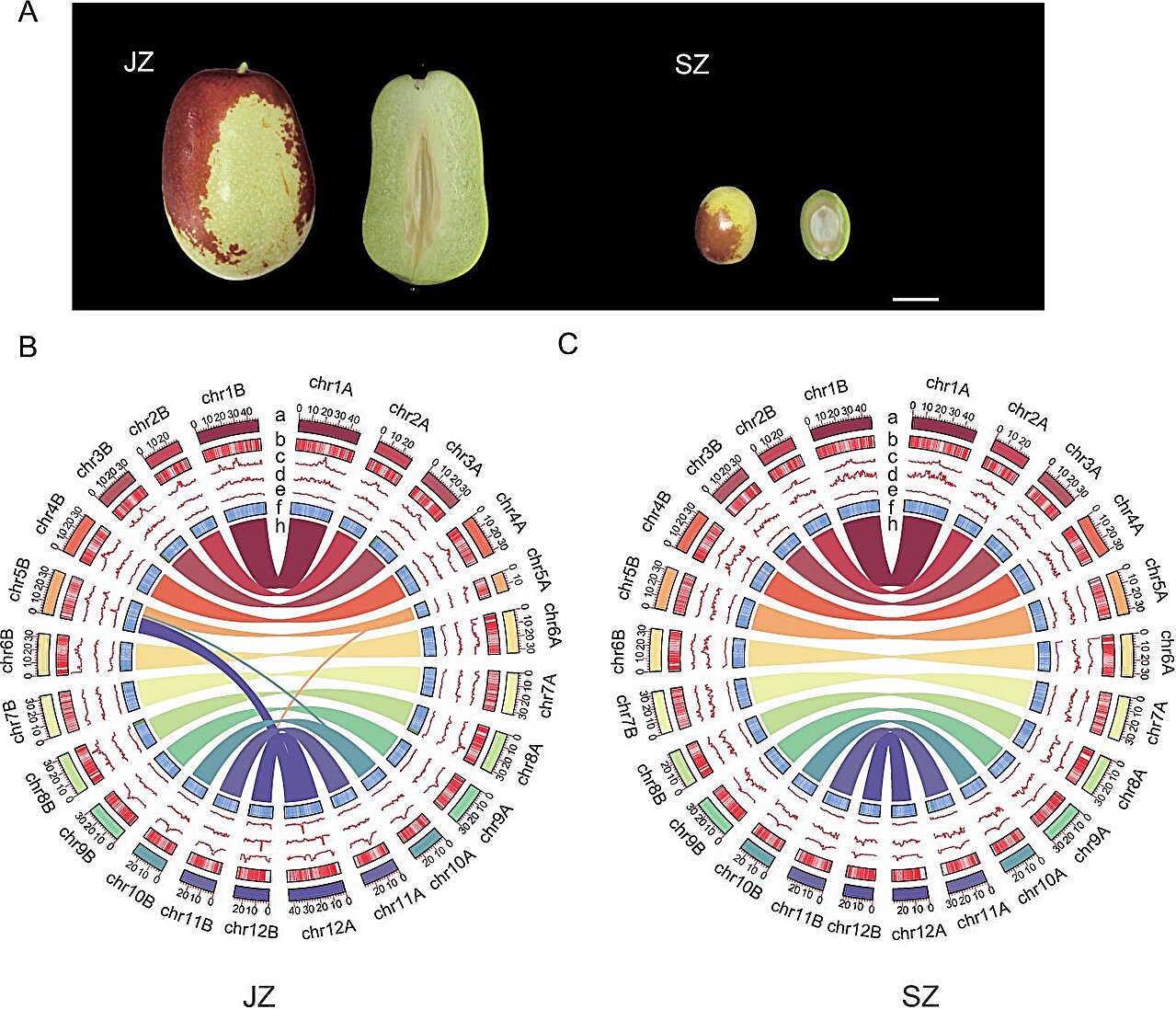
Chinese jujube, known for its economic and nutritional significance, was domesticated from its wild ancestor. While previous studies have shed light on some aspects of its domestication, many genetic details remain unexplored. The jujube’s transformation from a wild shrub with small, sour fruit to a cultivated tree with large, sweet fruit involved complex genetic changes.
Due to these unanswered questions and the economic importance of jujube, a comprehensive genomic study was essential to uncover the detailed genetic mechanisms underlying its domestication. This necessity drove researchers to delve deeper into jujube’s genetic evolution.
A team of researchers from Northwest A&F University and Xinjiang Academy of Agricultural Sciences have published a study on March 7, 2024, in Horticulture Research in which they report the assembly of haplotype-resolved genomes for the cultivated “Junzao” jujube and its wild ancestor. This research reveals significant genetic variations that have shaped jujube’s domestication.
The study successfully assembled the genomes of the cultivated jujube cultivar “Junzao” and its wild relative “Suanzao” using advanced sequencing technologies, achieving high-quality, haplotype-resolved assemblies.
These genomes revealed significant structural variations, including large-scale inversions and translocations on chromosomes 1, 3, 4, and 12. Comparative genomic analyses showed that these variations affect numerous genes, particularly those involved in starch and sucrose metabolism, which are crucial for fruit sweetness.
Additionally, the study identified new selection signals in regions associated with tissue development and pollination, highlighting the genetic pathways influenced by domestication.
These findings provide a detailed genetic map, offering valuable insights into the evolutionary and domestication processes of jujube. The comprehensive data not only enhance the understanding of jujube’s genetic diversity but also pave the way for targeted breeding strategies to improve fruit quality and other agronomic traits.
Dr. Jian Huang, a leading researcher, commented, “The haplotype-resolved genomes provide unprecedented clarity on the genetic underpinnings of jujube domestication. These high-quality genomes will be instrumental in breeding programs aimed at improving jujube fruit quality and resilience.”
The insights gained from this study have significant implications for jujube breeding and agriculture. The identified genetic markers and structural variations can guide targeted breeding strategies to enhance desirable traits such as sweetness and drought resistance. Furthermore, the comprehensive genomic data serve as a valuable resource for future research in jujube and other related fruit trees.
More information:
Kun Li et al, Haplotype-resolved T2T reference genomes for wild and domesticated accessions shed new insights into the domestication of jujube, Horticulture Research (2024). DOI: 10.1093/hr/uhae071
Citation:
From wild to sweet: Decoding the jujube’s genetic journey (2024, June 25)
retrieved 25 June 2024
from https://phys.org/news/2024-06-wild-sweet-decoding-jujube-genetic.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.