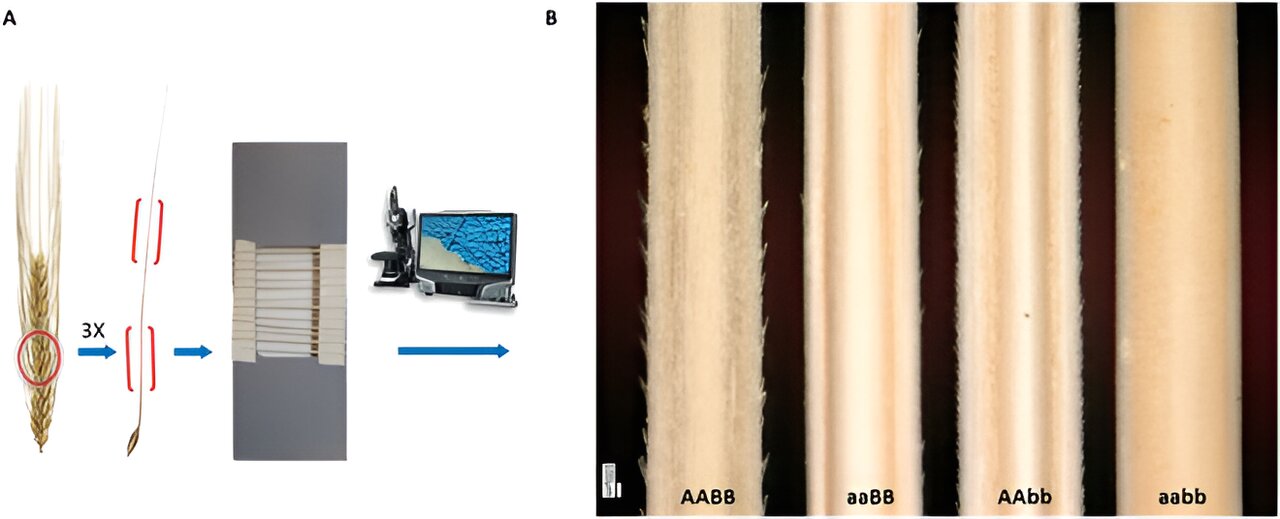
Awns, bristle-like extensions on grass crops like wheat and barley, are vital for protection and seed dispersal, with barbs on their surface playing a crucial role. While the genetic basis of barb formation has been explored through genome-wide association and genetic mapping, the detailed analysis of these small, variable structures poses a challenge.
Existing methods, such as scanning electron microscopy, provide detailed visualization but lack the automation required for high-throughput analysis. Therefore, the development of advanced automatic image processing algorithms, especially deep learning-based methods, to accurately segment and analyze the complex morphology of barbs is important for better understanding and growing cereal crops.
In August 2023, Plant Phenomics published a research article titled “Awn Image Analysis and Phenotyping Using BarbNet “.
In this study, researchers developed BarbNet, a specialized deep learning model designed for the automated detection and phenotyping of barbs in microscopic images of awns.
The training and validation of BarbNet involved 348 images, divided into training and validation subsets. These images represented diverse awn phenotypes with varying barb sizes and densities. The model’s performance was evaluated using binary cross-entropy loss and Dice Coefficient (DC), showing significant improvement over 75 epochs, with a peak validation DC of 0.91.
Further refinements to the U-net architecture, including modifications like batch normalization, exclusion of dropout layers, increased kernel size, and adjustments in model depth, led to the final BarbNet model.
This model outperformed both the original and other modified U-net models in barb segmentation tasks, achieving over 90% accuracy on unseen images.
The researchers then conducted a comparative analysis of automated segmentation results with manual (ground truth) data, revealing high conformity (86%) between BarbNet predictions and manual annotations, especially in predicting barb count. Additionally, the researchers explored genotypic-phenotypic classification, focusing on four major awn phenotypes linked to two genes controlling barb density and size.
Using features derived from BarbNet-segmented images, they achieved accurate clustering of phenotypes, reflecting the corresponding genotypes.
The study concludes that BarbNet is highly efficient, with a 90% accuracy rate in detecting various awn phenotypes. However, challenges remain in detecting tiny barbs and distinguishing densely packed barbs. The team suggests expanding the training set and exploring alternative CNN models for further improvements.
Overall, this approach marks a significant advancement in automated plant phenotyping, particularly for small organ detection like barbs, offering a robust tool for researchers in the field.
More information:
Narendra Narisetti et al, Awn Image Analysis and Phenotyping Using BarbNet, Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0081
Citation:
BarbNet: Awn phenotyping with advanced deep learning, potential applications in the automation of barley awns sorting (2023, December 28)
retrieved 28 December 2023
from https://phys.org/news/2023-12-barbnet-awn-phenotyping-advanced-deep.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.