Autophagy, a eukaryotic mechanism for breaking down cellular components, is a vital process in lytic organelles such as vacuoles in yeast and plants, and lysosomes in animals. Research has predominantly focused on the model plant Arabidopsis, revealing the conservation of autophagy-related genes across various plant species.
Recent studies highlight autophagy’s crucial role in plant responses to nutrient scarcity, stress management, and pathogen interactions, and its influence on plant yield and metabolism. However, while autophagy’s impact on lipid,primary and secondary metabolism in crops like rice and Arabidopsis is documented, its role in tomato cellular metabolism remains underexplored.
In June 2022, Horticulture Research published a research article titled “Autophagy modulates the metabolism and growth of tomato fruit during development.”
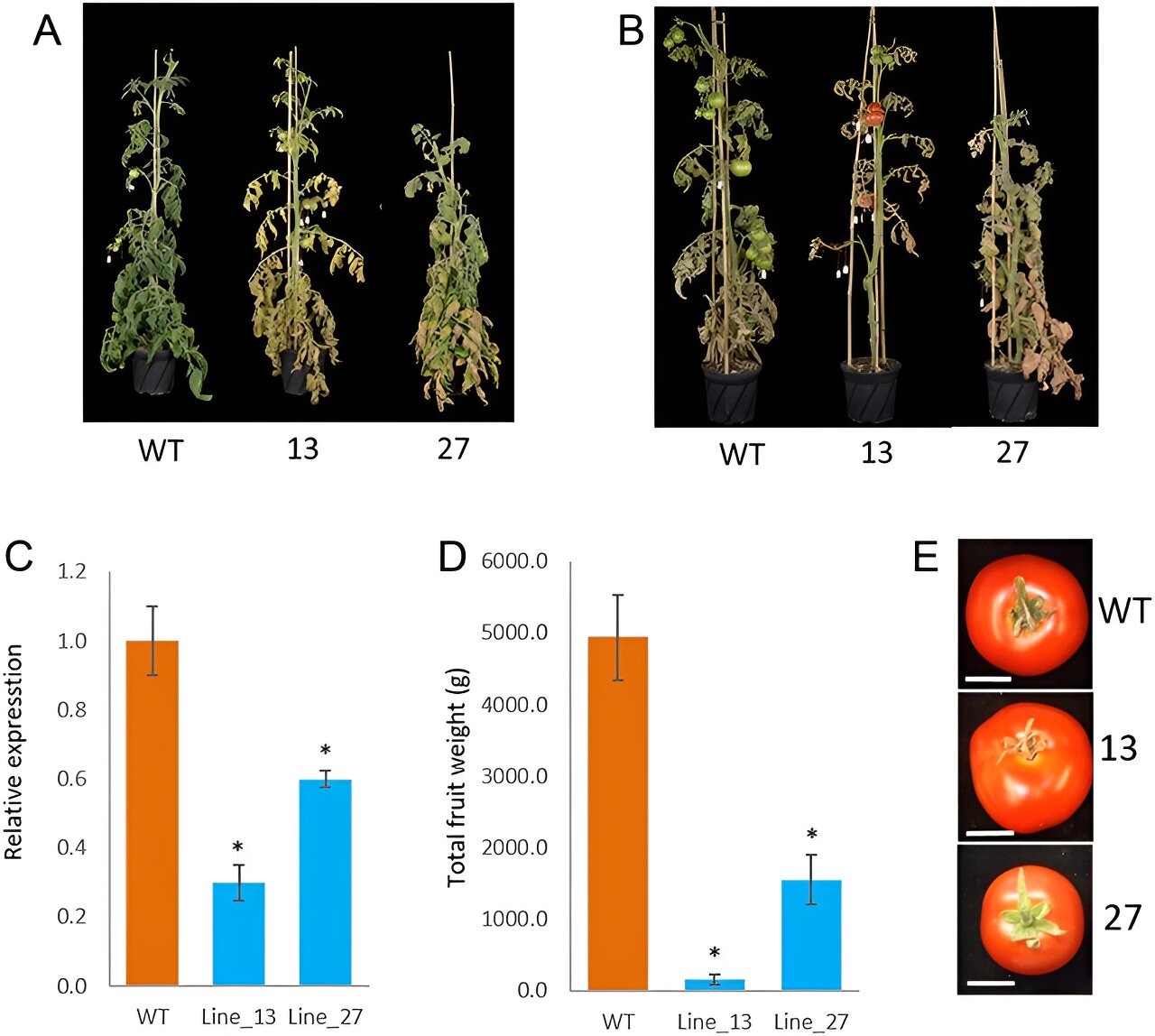
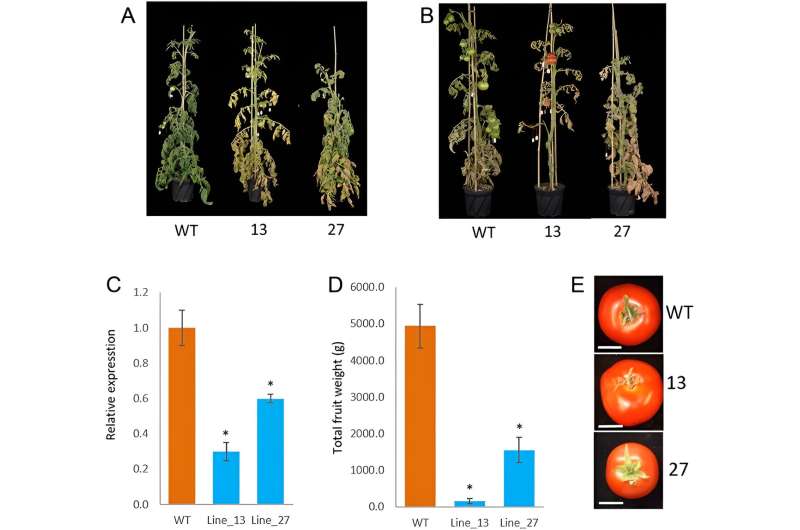
In this study, researchers employed RNA interference (RNAi) to specifically target and reduce the expression of the autophagy-regulating protease ATG4 gene. This approach led to the generation of tomato plants with distinct early senescence phenotypes and reduced fruit yields.
To analyze the effects of ATG4 downregulation, researchers conducted a series of experiments encompassing metabolite profiling, transcriptome analysis, and proteomics.
In ATG4-RNAi tomato leaves, only observed minor alterations in the levels of certain primary and secondary metabolites, such as citric acid, glycerate, and quercetin derivatives. However, in the fruit, more pronounced changes were evident. Primary metabolites like proline, tryptophan, and phenylalanine showed decreased levels in ATG4-RNAi lines, while secondary metabolites such as quinic acid and 3-trans-caffeoylquinic acid were substantially increased in ATG4-RNAi green fruits compared to wild type (WT).
Transcriptome analysis revealed significant upregulation of genes involved in organelle degradation and chloroplast vesiculation pathways in ATG4-RNAi lines. This was further corroborated by proteomics data, which indicated reduced levels of chloroplastic proteins.
These findings suggest that ATG4 plays a critical role in regulating chloroplast function and metabolism in tomato fruit. Additionally, lipid profiling demonstrated significant changes in lipid compounds in ATG4-RNAi lines, particularly at the green fruit stage.
A correlation network analysis of omics data pointed to strong associations between lipids, genes, and proteins involved in lipid metabolism, chlorophyll binding, and chloroplast biosynthesis. Finally, metabolic flux analysis using 14C- or 13C glucose in red ripe fruits showed decreasing rate of protein synthesis alongside the 14CO2 evolution on lack of autophagy in Arabidopsis.
However, there are few significant changes in the redistribution of metabolites, indicating that the absence of autophagy primarily affected specific metabolic pathways rather than causing widespread metabolic disruption.
In conclusion, this study reveals a key role for ATG4-dependent autophagy in tomato physiology. This is the first report linking autophagy to metabolic content in tomato pericarp, differing from previous studies focusing on foliar metabolism in Arabidopsis and maize.
This research provides a more comprehensive understanding of autophagy in tomato, and highlights the potential of targeting autophagy for future crop improvement and breeding strategies.
More information:
Saleh Alseekh et al, Autophagy modulates the metabolism and growth of tomato fruit during development, Horticulture Research (2022). DOI: 10.1093/hr/uhac129
Provided by
Plant Phenomics
Citation:
Unveiling the role of autophagy in metabolism and growth for tomato fruit development (2023, December 21)
retrieved 21 December 2023
from https://phys.org/news/2023-12-unveiling-role-autophagy-metabolism-growth.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.